Cross Institutional Bioimaging PhD Course
Level: PhD course
Time: September 2 to November 18, 2024
Locations: Copenhagen, Odense, Århus, Kng. Lyngby
Language: English
Number of contact hours: 80
Capacity limits: 20
Download and print the 2024 Cross-Uni-Bioimaging PhD Course Flyer
Course description
The Cross Institutional Bioimaging Ph.D. course will be given by a series of lecturers who are experts within each their field of bioimaging. The course will take place at different Danish research institutions in order to expose the students to different research groups, researchers and experimental research facilities. The course will thus give the students a unique opportunity of orienting themselves within an active and diverse field of interdisciplinary science within bioimaging.
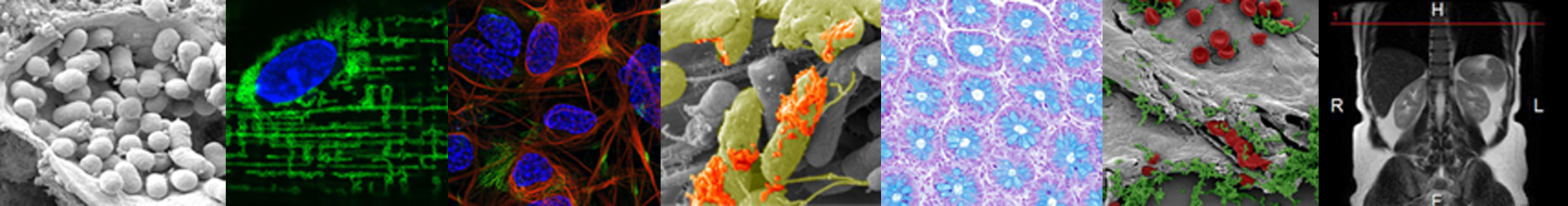
The course is relevant for PhD students within medicine, physics, chemistry, biochemistry, molecular biology, nano-bioscience, pharmaceutical sciences, agricultural science or biology. The emphasis of the course is a tour of all bioimaging techniques available in Denmark and will cover subjects like live cell imaging, confocal microscopy, electron microscopy, super-resolution microscopy, single particle techniques, stimulated emission depletion microscopy, imaging of neurons, cell migration and image analysis.
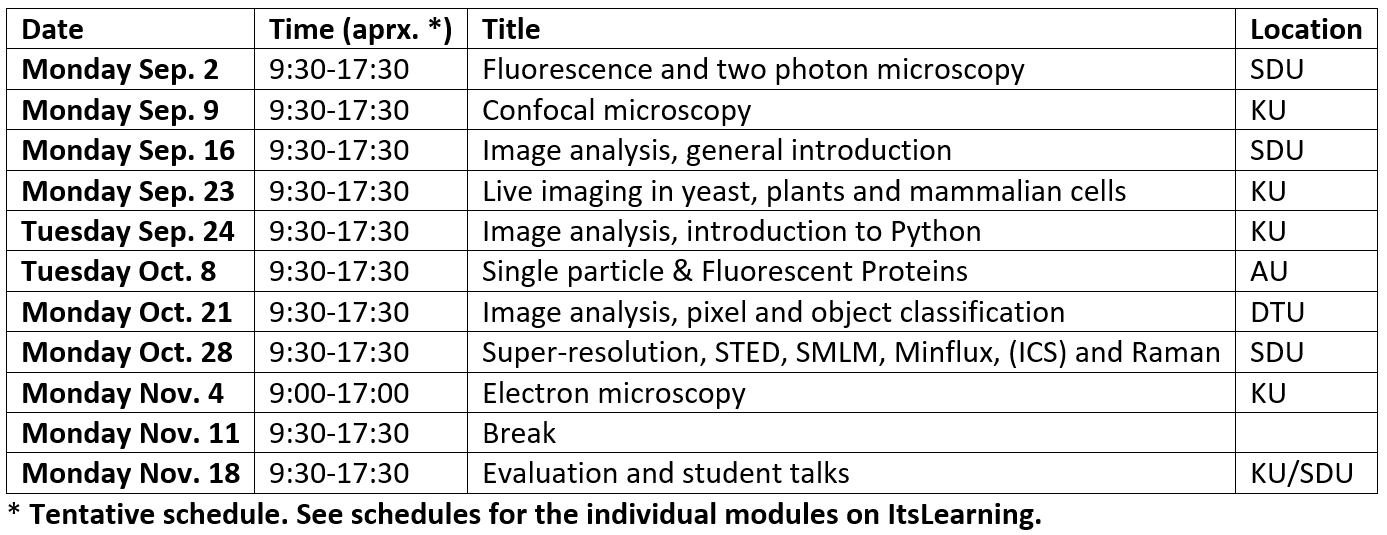
Time schedule for the course
The course runs consecutive Mondays for 10 weeks at 4 different universities and by different lecturers. Each module of the course covers around 8 hours, from 09:30-17:30. In general, the morning session will consist of a set of lectures and the afternoon session will predominantly involve either the student's active participation in experiments, specific numerical exercises, or inspection of the local experimental facilities.
Course credit and evaluation
The workload of the course corresponds to 10 ECTS points, the total workload includes reading material for each module and the preparation of the final talk (presentation). Credit for the course requires the student's presence at minimum 8 of 10 modules. For any missed modules, students will have to write a 3-page report on the topic of the missed module.
The student’s presence on the entire exam day is mandatory but the venue, KU or SDU, can be chosen by the student according to what is most convenient. The examination consists of 20 minutes of presentation and 15 minutes of questions from the examiners and student opponents. Each student must act as an opponent for the presentation of one other student. In the presentation each student will describe an experiment using one of the applications described in the course including an image analysis strategy. The task for the presentation will be given in advance (on the last module day the 8th of November).

Course information
Registration:
Registration can be requested by email to mfe@bmb.sdu.dk with information about your institution, department, supervisor(s) and your research topic.
Prerequisites:
Students must hold a master’s degree in physics, engineering, life science, biology or medicine and accepted in a PhD program.
Course responsible:
Jonathan Brewer (SDU)
Lecturers:
(AU) Victoria Birkedal, Mette Galsgaard Malle, Thomas Boesen
(DTU) Rasmus Reinhold Paulsen, Anders Bjorholm Dahl, Paul Kempen
(KU) Clara Prats, Ivana Novak, Michael Lisby, Alexander Schulz, Jon Sporring, Klaus Qvortrup
(SDU) Eva Arnspang Christensen, Martin Hedegaard, Jonathan Brewer, Daniel Wüstner, Helge Thisgaard
Literature:
Will be announced every week during the course or be found on the course page on the ItsLearning platform.
Course homepage:
General course information is found on this current page (https://dambic.dk/index.php?page=Cross-Institutional-Bioimaging)
Course materials and more detailed course plans will be published on the ItsLearning platform (SDU).
Questions? Please contact Morten Frendø Ebbesen: mfe@bmb.sdu.dk

Short description of course modules
Fluorescence and two-photon microscopy
Jonathan Brewer
This day will cover the basics of fluorescence and epifluorescence widefield microscopy which form the bases for understanding many of the more advanced techniques introduced later in the course. This will be followed by an introduction to multiphoton excitation microscopy. A session with flash-presentations from all participants will also be included in the program to facilitate interactions between the participants.
Confocal microscopy
Clara Prats
Students will be introduced to the Principles and Essentials of Single Point Scanning Microscopy. Lectures and hands-on practical exercises will be combined to teach the students the critical components of a Confocal Microscope and, how to properly construct imaging light paths and settings to avoid artifacts and collect proper bioimaging data.
Image analysis, general introduction
Daniel Wüstner
In this module, students will be introduced to image processing techniques commonly used in bioimage analysis from a theoretical and practical perspective. The first theoretical part introduces to image formats, signal-to-noise, image depth, contrast, look-up tables, use of histograms and convolutions . The second theoretical part will focus on image stacks, image segmentation and batch processing. The third theoretical part introduces analysis of live-cell imaging data. The practical part consists of hands-on exercises of selected topics using the GUI of ImageJ, co-localization analysis and self-developed ImageJ Macro scripts.
Live imaging in yeast, plants and mammalian cells
Ivana Novak, Michael Lisby, Alexander Schultz
Joint morning lectures for all students will be followed by three separate demonstrations/hands-on of various techniques in live cell imaging in yeast, plants and mammalian cells. Students should choose one demonstration to follow.
Alexander Schulz, Frederiksberg Campus
Live cell imaging of plants is able to uncover development-related physiological changes and responses due to abiotic and biotic stress. We will present examples including cell signalling, the cell-to-cell and long-distance transport of photoassimilates, and the defence of plants against pathogens. Show-cases at the point and spinning confocal microscopes will present the opportunities and limitations of advanced microscopy approaches in this research field.
Jakob Grunnet Knudsen & Nynne Christensen, North Campus
Live cell imaging of mammalian cancer cells or endocrine cells will be demonstrating fast responses at the plasma cell membranes, cellular signalling and signalling between cells. Show-cases of CLSM, FRET, video rate imaging and long-term imaging will be presented.
Michael Lisby, North Campus
Budding yeast Saccharomyces cerevisiae is a widely used eukaryotic model for evolutionarily conserved processes. We will show-case an example of how specific protein-protein interactions can be monitored during the DNA damage response using Bimolecular Fluorescence Complementation (BiFC) on a high-resolution fluorescence wide-field microscope.
Image analysis - introduction to Python
Jon Sporring
In this part of the course, you will get a quick introduction to image processing using Python. Using your own computer, you will learn how to do write short programs that perform simple noise reduction, segmentation, and object analysis on 2 dimensional images. We will work with example images, and you are welcome to bring your own as further work cases. Before class, you are expected to have installed Anaconda and have basic familiarity with the Jupyter notebook. If you have no programming experience in python, then it is strongly recommend that you work through the Introduction to Python Programming material to be distributed prior to class.
Single molecule fluorescence
Mette Galsgaard Malle, Victoria Birkedal
The first part of this module will give an introduction to single molecule fluorescence and describe fluorescence microscopy with high signal-to-noise ratio and spatial resolution for single molecule studies. This will include the theory of total internal reflection (TIRF) microscopy. We will present the principles and selected applications of single molecule FRET spectroscopy.
The second part will focus on real time imaging with TIRF microscopy and present application examples, including how single liposome imaging can constitute a parallel screening platform with ultra high sensitivity. This will also include an exercise with single molecule data.
Besides lectures and exercises, the module will include a lab tour.
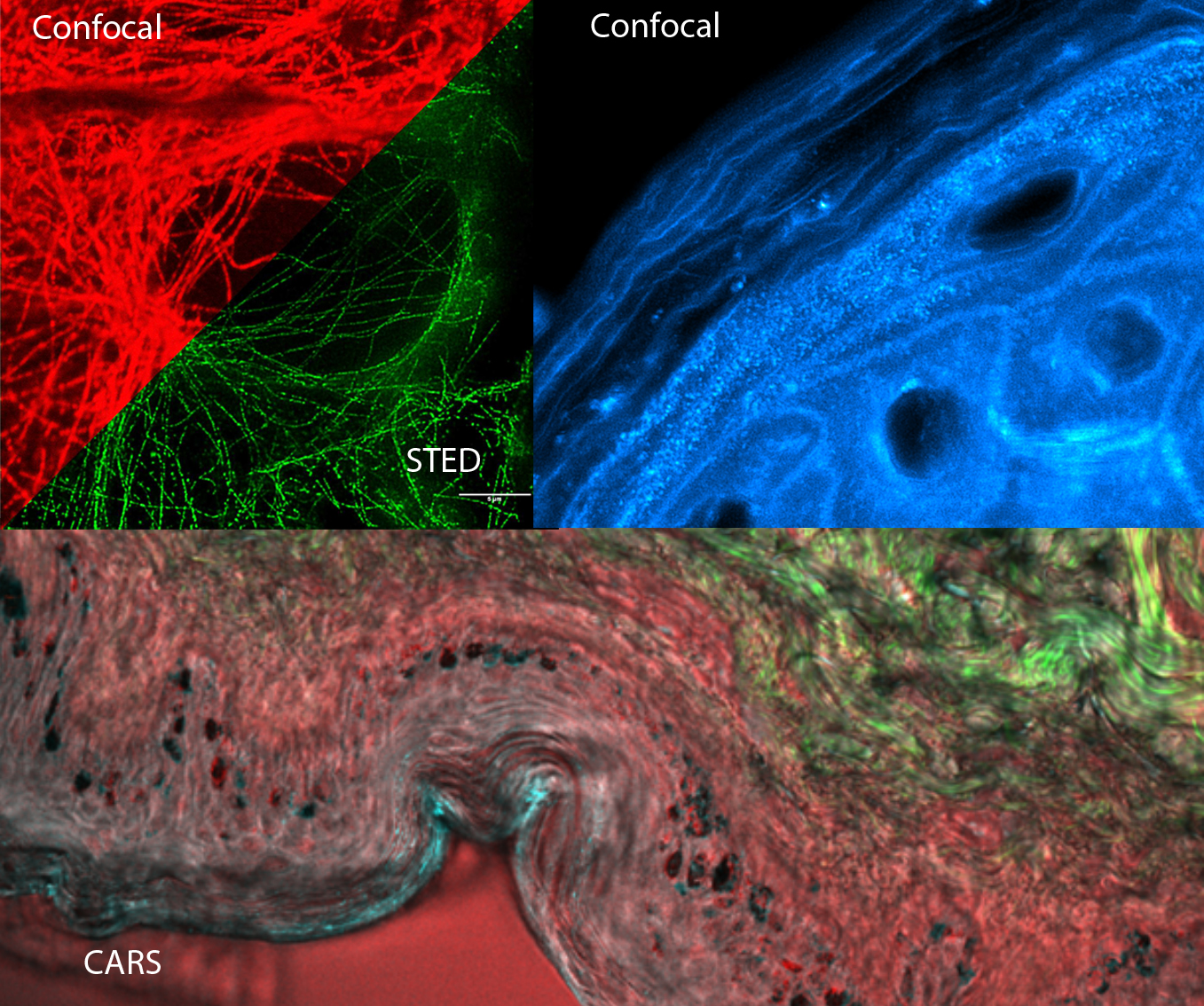
Super-resolution, STED, ICS and Raman
Eva Arnspang Christensen, Jonathan Brewer & Martin Hedegaard
This day will cover a wide range of advanced imaging techniques. Specifically, the super resolution techniques including stimulated emission depletion microscopy (STED), structured illumination microscopy (SIM) and localization-based microscopies such as photoactivatable localization microscopy (PALM) will be introduced.
Coherent anti-Stokes Raman scattering (CARS) and confocal Raman imaging will be presented. They are label-free imaging technique based entirely on molecular vibrations. This course will include an introduction to basic working principles and choice of instrumentation including lasers, microscopes and spectrometers. In addition, there will be an introduction to pre-processing and analysis of Raman imaging data.
Furthermore, methods for dynamic measurements in samples such as fluorescence correlation spectroscopy (FCS) and k-space image correlation spectroscopy (kICS) will be shown. These techniques facilitate measurements of bulk diffusion coefficient from conventional confocal, epi fluorescence or TIRF imaging.
Image Analysis - pixel and object classification
Rasmus Reinhold Paulsen, Anders Bjorholm Dahl
The image analysis course at DTU Compute is focused on two topics that are relevant for biomedical image analysis. The first topic is pixel classification, where the goal is to assign a relevant label to a given pixel. Labels are normally predefined like for example background, nuclei, and membrane. The classification is based on a using a training set of images to create a statistical assumption on the distribution of pixel values within the classes.
The second topic is BLOB analysis, where the goal is to classify objects in an image based on their shape. Typical examples are nuclei detection in cell images or organ identification in medical scans.
Non-invasive imaging modalities (PET-SPECT-CT-R) (not in 2024)
Helge Thisgaard
The course will cover the basics concepts of preclinical nuclear medicine techniques using PET/SPECT/CT imaging modalities. The theory behind the techniques will be presented followed by hands-on experience in the laboratory
Electron Microscopy
Klaus Qvortrup
The course provides an introduction to the essential grounding in the basic principles of electron microscopy, covering topics such as electron optics, electromagnetic lenses, principles of transmission and scanning electron microscopy, electron sources, vacuum systems, specimen-electron interactions and diffraction. The state-of-the-art facilities available at CFIM allow for a strong practical element of demonstrations of both cryo- and room temperature electron microscopy. The course will be run by experienced microscopists in a relaxed atmosphere with the aim of promoting discussion and exchange of ideas between students and tutors.
